The analysis pipeline¶
Although the basic pipeline for analyzing whole exome sequencing, as propuested in NCBI, consists of three basic phases (secuencing retrieval, aliengment, and variant calling), more steps are distinguished to make the pipeline more flexible:
Secuence retrieval
Reference genome retrieval
Secuence alignment
Alignment processing
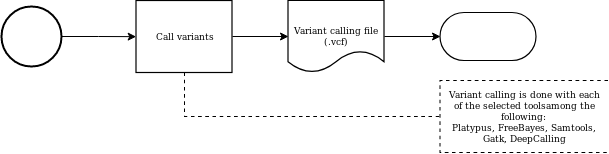
Variant calling
Variant calling evaluation
Secuence retrieval¶
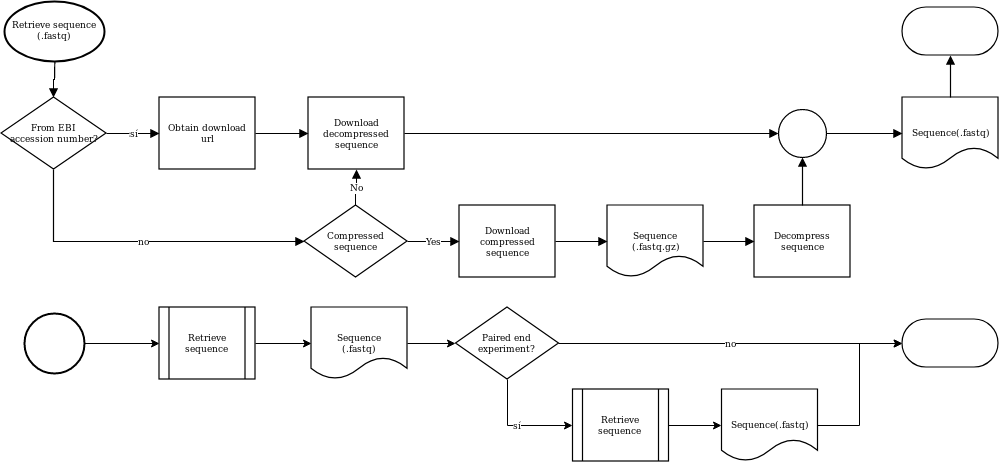
Retrieving the secuencing is a step that takes into account wether the experiment is paired end, or the sources are gz compressed.
Reference genome retrieval¶
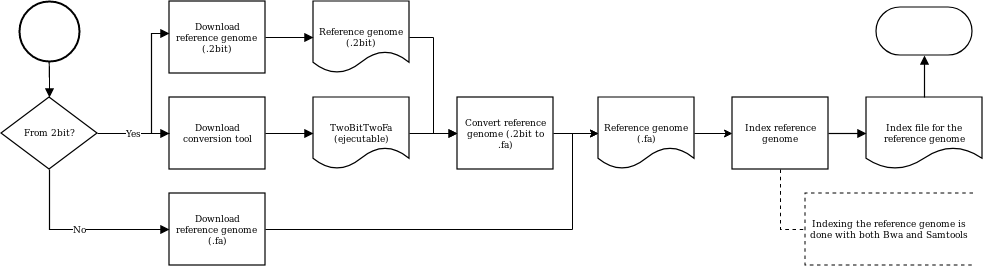
The reference genome, essential for more steps than the alignment, may be obtained in 2bit format (and then converted to fa).
Secuence alignment¶
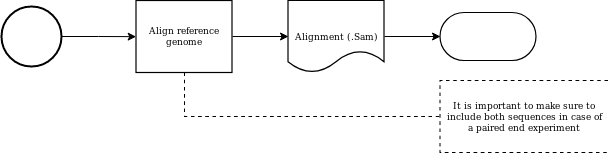
Aligning the secuencing against the reference genome is a process that produces a not sorted, nor indexed, sam file.
Alignment processing¶
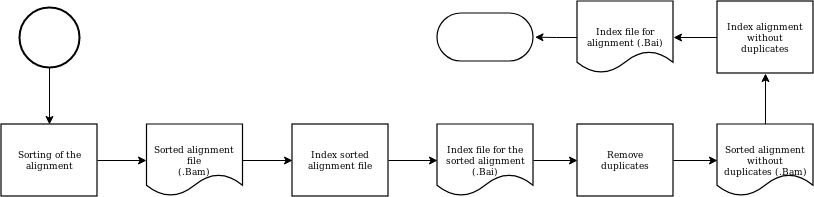
Processing the alignment includes sorting, indexing and removing duplicates from the original alignment. In the end, it produces a bam and bai file.
Variant calling evaluation¶
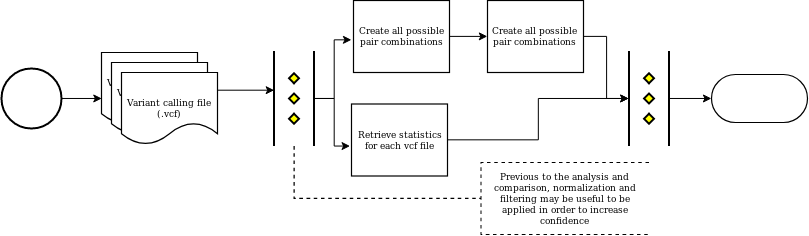
Variant calling comparation and estatistical summaries for the variants identifyed.